Conda
Most of the tools need some requirements. Some need lot of requirements: Python or R libraries in specific version, the last brand new compilator, or simply a newer one. Often, those dependencies are not compatible with what we already have on our system or between them.
Conda is an open source package, dependency and environment manager for any language: Python, R, Ruby, Lua, Scala, Java, Javascript, C/ C++, FORTRAN. Miniconda is a small “bootstrap”
version that includes only conda, Python, and the packages they depend on. Over 720 scientific packages and their dependencies can be installed individually from the Continuum repository
with the conda install command.
This allow us to install tools within some dedicated and isolated environments.
"Private" Conda environments
We don't recommend installing tools on your own if the required tool is already available as a Conda package in the Bioconda or Conda-Forge channels. Conda is already installed on the cluster — you just have to activate it for your user.
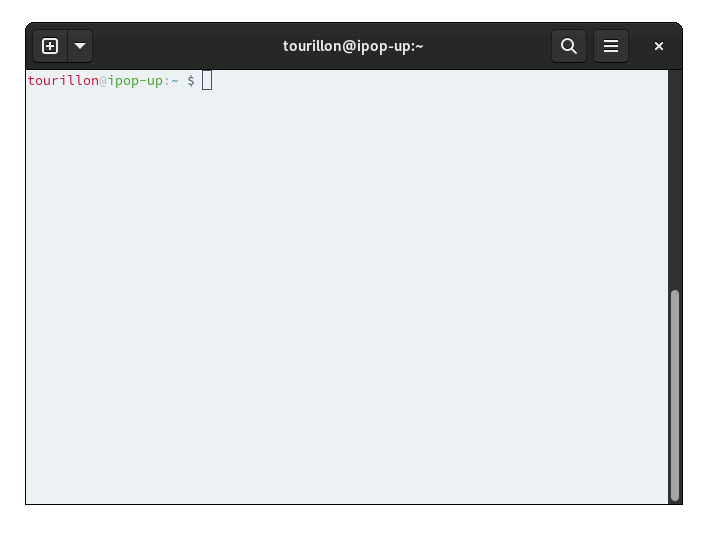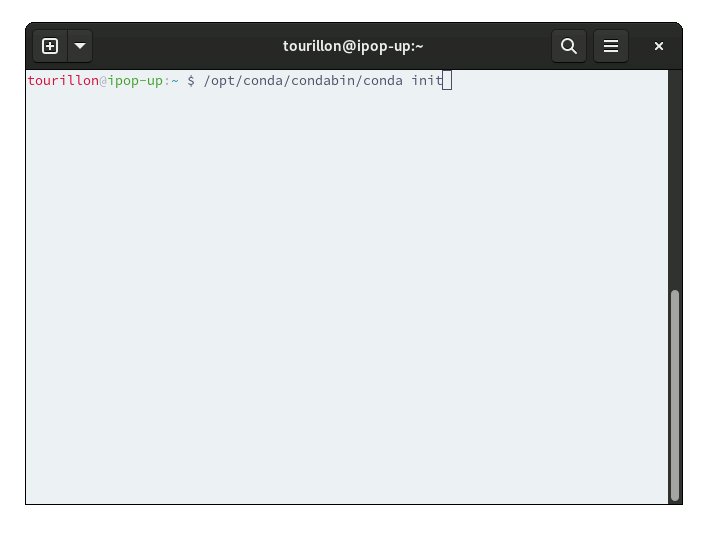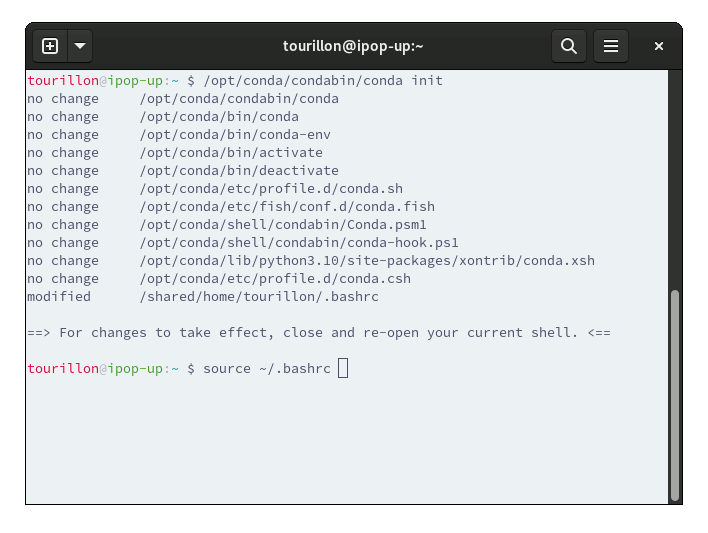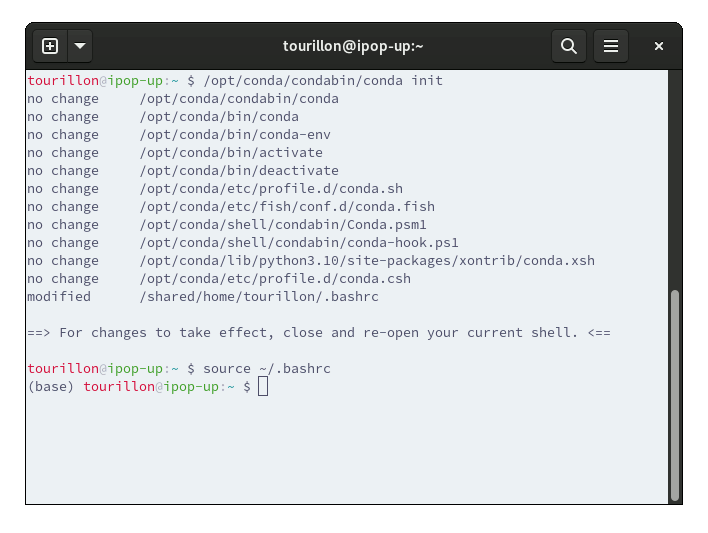
You can initialize conda with the following command :
/opt/conda/bin/conda init
source ~/.bashrc
Your home directory isn't designed to store lots of files. If you really need to install Conda packages, please install them in your project directory:
# create a virtual environement named 'my_env' in '/shared/projects/<project_name>/conda/envs/'
conda create -p /shared/projects/<project_name>/conda/envs/my_env
💡 Alternatively
You can configure Conda to automatically install environments and packages in your project directory by editing the configuration file
~/.condarc:Edit with:
bash vi ~/.condarcExample content:
yml envs_dirs: - /shared/projects/<project_name>/conda/envs pkgs_dirs: - /shared/projects/<project_name>/conda/pkgs
"Shared" Conda environments
Those environments are provided by the IFB, you can load them using the module command as explain in this page.
If you need it, you can ask us for a shared environment. There are different use cases:
- I don't know how to use Conda,
- I'm preparing a training session and I want that all the attendees have the same software environment,
- Conda packages can be heavy in term of disk usage.
To request a tool or a Conda environment, 2 solutions:
- Propose one via our dedicated git repository cluster/tools
- Request a tool on our community forum: https://community.france-bioinformatique.fr/
To know if a package and a specific version is available in the channels bioconda and conda-forge:
conda search -c conda-forge -c bioconda mu_tool
Cons of Conda
Because those environments isolate the software, other Python, R or Perl libraries installed on the system or on other Conda environments are not available within the Conda environment. If it is an issue for you, please let us know.